Technologies
4th generation sequencing technique innovated by Genvida.
SONAS
(SOlid State NAnopore Sequencing)
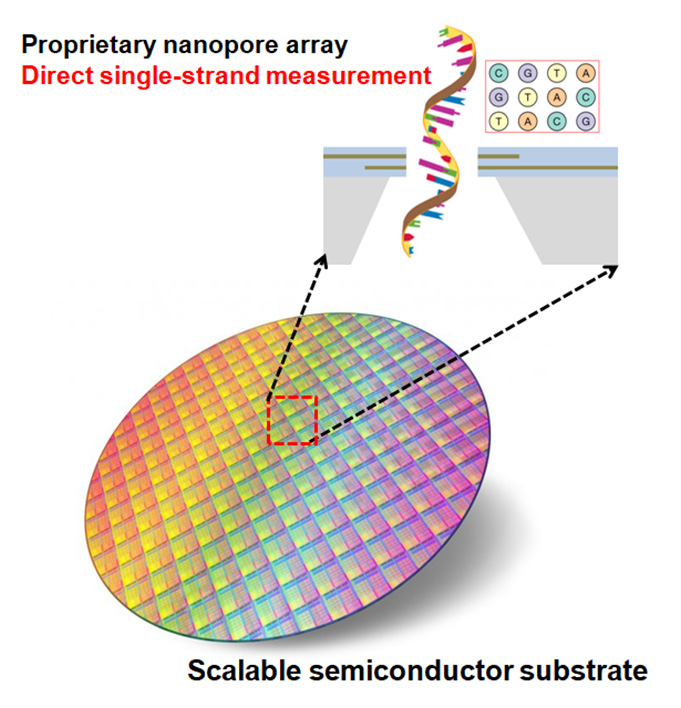
SONAS makes use of proprietary technique to achieve massive and parallel fabrication of nanopore arrays with pore size down to <5 nm in one shot. Since the fabrication involves only semiconductor manufacturing compatible processes which are highly reproducibility with nanometer scale accuracy, the nanopore array fabrication can be easily scaled up for wafer-scale mass production.
Combining with the uniquely developed sample preparation technique (gPrep) and integrated bioinformatics suite, SONAS offers a drastic improvement over the competing technologies. Advantageously, Genvida’s direct read SONAS technology permits exceptionally high sensitivity at single molecule level rendering PCR or other types of amplification unnecessary, hence simplified workflows; moreover, uniquely because of the optimal combination of mean read length and unprecedentedly high throughput, SONAS makes the lowest possible per base and per run cost, and with the assurance of high quality results by our systems’ ultrahigh accuracy (>99.9% per base, i.e. an order of magnitude better comparing to the current best).
As a low cost, real time sequencing system with high accuracy, high throughput, fast sequencing speed and long read length is required to unlock the potential market, which entails a drastic change in the current sequencing approach and a quantum leap in sequencing technology. With the “all physical processes” approach adopted by SONAS, Genvida possesses a definite competitive edge to knock out current technologies. This will enable a much wider market than that of the current technologies. Data driven, informed decisions can be made by the physicians, the government officials and, ultimately, the patients themselves.
Built upon a set of common core patented technologies, our platform patented technology SONAS® can germinate into different families of high value, high growth products and applications, such as genome mapping, digital PCR and single cell systems, which will be developed at a later stage for potential business expansion in the future.
Fundamentally, our SONAS® based sequencing platform overhauls the underlying operating principle of the aforementioned ‘shotgun sequencing’ by offering an easy to use portable sequencing system which includes gPrep: a simple sample preparation procedure/protocol, gSeq(a single-use disposable biochip and multi-series devices with SONAS® inside), gCal(an integrated portable DNA sequence analyzer with artificial intelligence calculation ability), and gInfo(an initiative and powerful bioinformatic software suite).
Our Platform
gPrep
Instead of randomly cut the nucleic acids by ultrasound to create the short oligos for shotgun sequencing, we make use of the endonucleases to systematically cut the nucleic acids according to the restriction sites. The restriction sites will serve as markers and will greatly simplify the assembly and mapping of the reads during the post-detection analysis. Adaptable sample preparation protocols compatible to many NGS technologies.
gSeq
gSeq is a product series including chip for DNA sequencing, and detection circuitry together with microfluidics channel. All products are built base on SONAS® platform technology and embedded with an array of solid-state nanopores. In each nanopore, there are embedded electrodes, from which the physics based nucleotide detections are performed.
gCal

This portable unit serves as the command and control of the gSeq chip or devices. It is the interface for the application platform of SONAS®, and functions as the exchange media between the operator and cloud server. Data from the gSeq devices will be stored and analyzed through artificial intelligence for DNA sequence characterization. Also, the data involving the whole sequencing process will transmit to cloud server in real-time.
gInfo

It is an integrated bioinformatics suite allowing rapid field diagnostics. Functioning together with gSeq devices and cloud server, it works in data analyzing and DNA sequence reporting. Moreover, gInfo acts as information exchange platform with other universities, research institutes and hospitals through collaboration for the sharing of cloud databases. The resulting bioinformatics will be effective for exploring numerous applications including bio-screening, bio-surveillance, gene profiling, development of personalized medicines etc.









